
Plugins
Plugins have been introduced recently to Minerva and allow the development of user-specific tools to explore and analyze the map or to provide connections to other databases.
They are loaded through an URL to their compiled .js file as shown in the following figure. Plugins that are mentioned below are already suggested and can be loaded directly with one click.
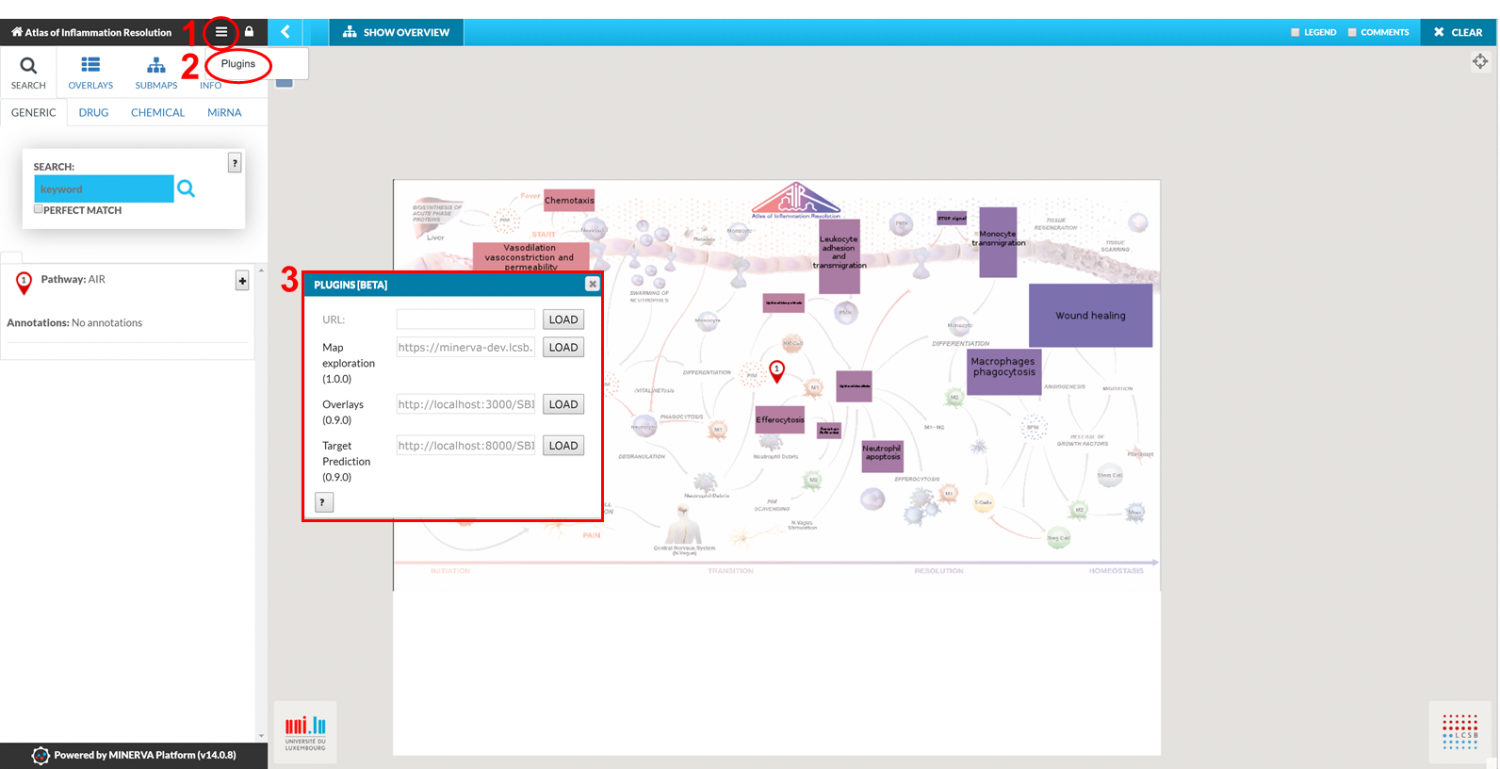
AIR Plugins for Disease Maps analysis:
The "AirPlugins"-Plugin consists of two major parts, each providing many functionalities to make use of the data stored in the AIR and generate new hypotheses.
- Xplore: Data-independent exploration of the AIR. Search interactions, export data, visualize network topology features, and find regulators of selected phenotypes.
- Omics: Analysis of -omics data provided by the user. Estimate phenotype levels and predict key regulators in the samples.
Additionally, more sub-plugin will be developed in the future, allowing the integration of more data types into the AirOmics analyses:
- Variant: Mapping of genomic variant data and prediction of variant effects.
Additional resources:
Using AirPlugins on other Disease maps
With our research, we aim to support open-source science engagements. Therefore, all information on the AIR, either submaps, phenotypes sets, or JavaScript code of the plugins, is publicly available. To use our plugins on other disease maps than the AIR, the javascript and data files need to be copied and pasted to a new directory that is publicly accessible by MINERVA, for example on GitHub. In the AirPlugins.js file, the variables "URL_PLUGINFILES" and "URL_DATAFILES" need to be edited with the new URLs in a text editor. Now the URL for the raw code of the AirPlugins file (which can be renamed) can be loaded in MINERVA.
However, the data in the plugin is not fetched from the MINERVA project in which they are loaded, but on the information in the public data files. Two data files are required:
- Elements.json: A JSON dictionary describing properties of elements defined by a string ID
- Interactions.json: A JSON list of dictionaries describing interactions between a source and target element
Both files require several attribute values, some of which are specific for the unique design of the AIR and can be irrelevant for other projects. To generate the data files for their MINERVA projects, users have to contact the AIR developing team.
Other plugins developed by the AIR team:
These plugins are usable on all MINERVA projects.
- Overlays: automatically create, display or remove multiple overlays from uploaded data files
https://raw.githubusercontent.com/sbi-rostock/AIR/master/PlugIns/Overlays.js - UserAccount: allows the users to edit their Minerva user profile information
https://raw.githubusercontent.com/sbi-rostock/AIR/master/PlugIns/UserDetails.js - Centrality: calculate network topology values independent of the Minerva project for all interactions or a specific map
https://raw.githubusercontent.com/sbi-rostock/AIR/master/PlugIns/Centrality.js
Plugins developed by the MINERVA team:
(see Minerva documentation on plugins)
- Disease variant associations: Provide information about genes associated with the given disease in the context of the selected disease map.
- Drug reactions: Explore the adverse reactions of the drugs which are interacting with the presented entities in a disease map.
- Exploration: Enhance the visualization and exploration of molecular interaction in the map.
- Guide: Provide an introductory guide through various aspects of the MINERVA platform.
- GSEA: Calculate the enrichment of the elements from the selected overlay in the pathway.